Research
A list of my current, past research interests, and projects that I am working upon!
Current Research

Genomic Dependencies in Cancer
Cancer evolution is driven by the emergence and selection of genomic alterations. In a growing tumor, proliferating cancer cells accumulate alterations over time, some of which modify the tumor phenotype by conferring advantageous features. In evolutionary terms, these alterations are selected. Understanding which alterations are selected and in which contexts do they provide an advantage are urgent questions in cancer genomics. When two alterations influence each other likelihood of being selected, we say they are evolutionary dependent. Currently working on expanding the previous work done by (Mina et al).
Checkout the (SelectSim) package developed for the same.
Previous Research Topics
-
Integrative Analysis and Machine Learning based Characterization of Single Circulating Tumor Cells
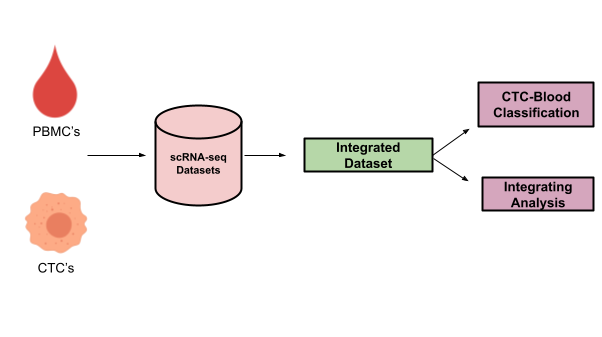
Cancer cells, after detaching from solid tumors migrate through the bloodstream to colonize at distant organs, leading to the development of cancer metastases. Cancer cells under circulation are called circulating tumor cells (CTCs). I worked on the project of integrative analysis of CTCs under Prof Debarka Sengupta (lab) where we collated publicly available single-cell expression profiles of circulating tumor cells (CTCs) and showed that CTCs across cancers lie on a near-perfect continuum of epithelial to mesenchymal (EMT) transition. Integrative analysis of CTC transcriptomes also highlighted the inverse gene expression pattern between PD-L1 and MHC, which is implicated in cancer immunotherapy. More information about the work is published and can be read (here)
-
Search of scRNA-seq profiles using GPU
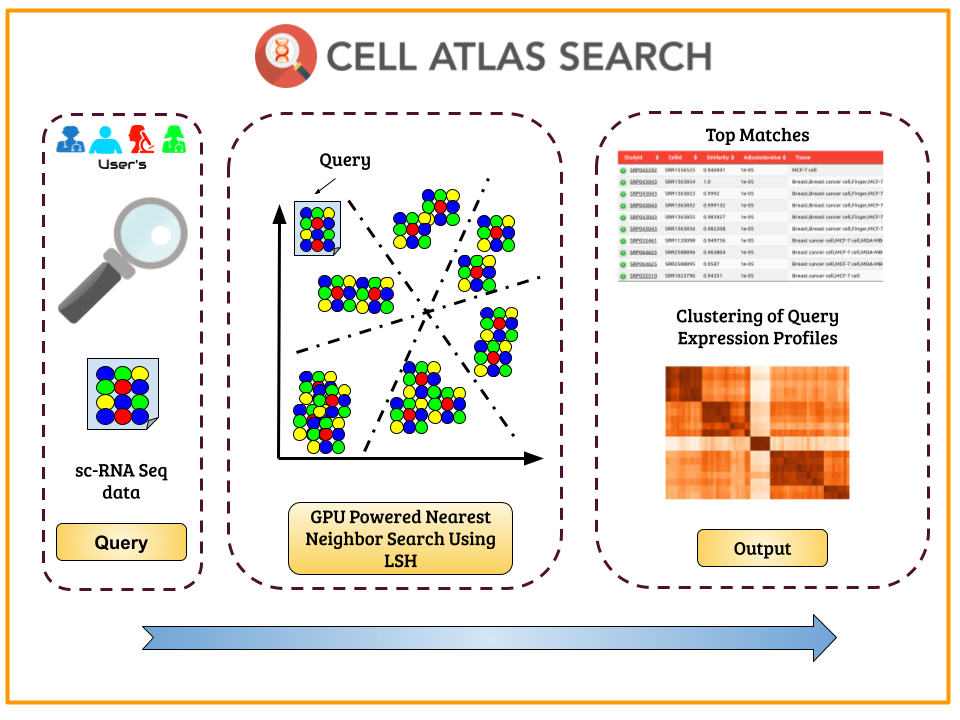
An explosion in production of single-cell expression data has triggered the need for a search engine. To cater to the need of the hour, we developed CellAtlasSearch, a novel search architecture for high dimensional expression data, which is massively parallel as well as light-weight, thus infinitely scalable. In CellAtlasSearch, we use a Graphical Processing Unit (GPU) friendly version of Locality Sensitive Hashing (LSH) for unmatched speedup in data processing and query. Currently, CellAtlasSearch features over 300 000 reference expression profiles including both bulk and single-cell data. I worked on this project under Prof Debarka Sengupta (lab). The server can be explored at this (link) and paper can be read (here)
-
Revealing Dynamic Architecture of Lipidated Proteins
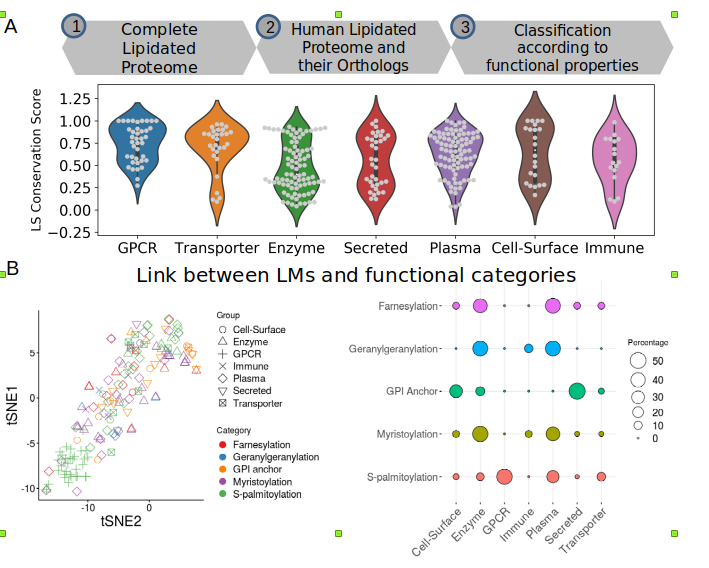
Proteins Post-Translational Modifications (PTMs) act as a multi-layered regulation mechanism for selective and controlled expression of cellular proteins. Lipidated proteins are one of the major components of virtually all cells, whereby lipid chains are attached covalently to the target residue site. We performed a proteome-scale integrated analysis on ∼10,000 unique lipidated proteins using publicly available repositories and identified >3,00,000 protein orthologs. The results of this comprehensive study show that location of lipid modification site is rapidly evolving and, is highly coupled with protein function and location. We also build a neural network classifier to predict lipidation site. I worked on this project as my Master thesis under Prof Lipi Thukral (lab) and Prof Angshul Majumdar (lab)
